Among the memorable talks was the one from Juan Luis Cano Rodríguez on Jupyter (see abstract) and the role of IPython notebooks for reproducible science. With all his energy and love of Python, this little guy will fly far. The key idea behind the notebooks is to display and run your code in the browser, embedding the output in the page, be it simple numbers or complex graphics. When the Python (or R/Julia/...) kernel is shut down, the page becomes frozen and ready to be shared. Others can quickly browse it or re-run the code as they wish. Just provide a link to download the input data and you have the perfect mix for reproducible experiments. See here for a live demo in the Nature journal.
Juan starting his talk on Jupyter.
The second talk which just made my day is Romain Guillebert's presentation on Pypy (see abstract), an alternative implementation to CPython, and pymetabiosis, a bridge between CPython and Pypy. He ran a Sobel edge detection coded in pure Python on images stored as python lists: roughly speaking, CPython was running at 2 seconds per frame and Pypy at 25 frame per seconds. To remove any doubt, he did it both with a recorded video and a live feed from his webcam. If all it takes is
s/python/pypy/, then I take it: it is much less code adaptation than all the alternatives proposed in the high performance talks (e.g. numpy, numexpr, numba and cython). Guido Van Rossum mentioned it when he came to Imperial: if Python is slow, use Pypy. Romain did a perfect demonstration of it. Of course, Pypy is not ready yet. But shall we switch to a new and faster language where image processing packages might be non-existent or in their infancy, such as Julia, or shall we instead spend our efforts making Python faster? Furthermore, there are companies interested in a faster Python implementation, such as Lyst (where Romain works) who are giving a try at Pypy, or Dropbox (where Guido works), who are working on Pyston, their own implementation of Python. There is the cost of porting existing code, and is it worth losing the simplicity of the Python syntax? Be it Julia, Pypy or others, the global switch will happen when a certain critical mass is reached, both in term of user community and maturity of key packages. It is a good time to choose a side.
Romain during his talk.
At the end of my talk on detecting the organs of the fetus in MRI data using Python, I have been asked: if you had to do it again, would you still have chosen Python? My answer is yes, because of all the existing libraries and their communities (in particular scikit-learn and scikit-image), the simplicity of the Python code to express complex ideas, and the ability to painlessly interface it with C++ through Cython. But could I have used Pypy?
Now let's get started with Pypy.

Before rushing into it, you might want to check the list of packages that are known to be compatible. Anyway, it is the beauty of open source, if it does not suit you out-of-the-box, just fork it, fix it and share it.
wget https://bitbucket.org/pypy/pypy/downloads/pypy-2.6.0-linux64.tar.bz2 tar -xvjf pypy-2.6.0-linux64.tar.bz2
Create an alias to the pypy binary and try it. If it cannot find installed Python packages, edit your PYTHONPATH to include obvious paths such as
/usr/lib/python2.7/dist-packages/ , or wherever those packages are installed in your CPython distribution. If who wonder where a package is installed, run pydoc package_name and look for the location of the main file at the top of the doc.If you don't have it yet, get pip:
easy_install --install-dir=~/.local/ pip
The next step is to install numpy.
git clone https://bitbucket.org/pypy/numpy.git cd numpy pypy setup.py install --user(--user for local install in ~/.local)
and pillow ("the friendly PIL fork") to write images, and joblib for easy parallelisation:
pypy -m pip install setuptools pillow joblib
Now let's do an experiment: I want to play with Random Forests using customised weak learners, I wrote simple Python code which does exactly what I want, but is quite slow, let's see what adjustments to the code Pypy require, and the speed gain.
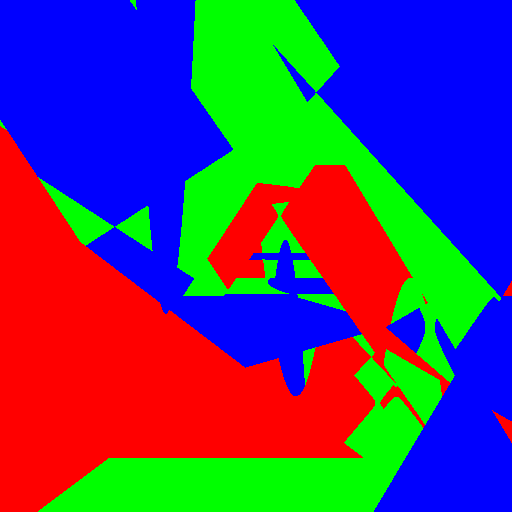
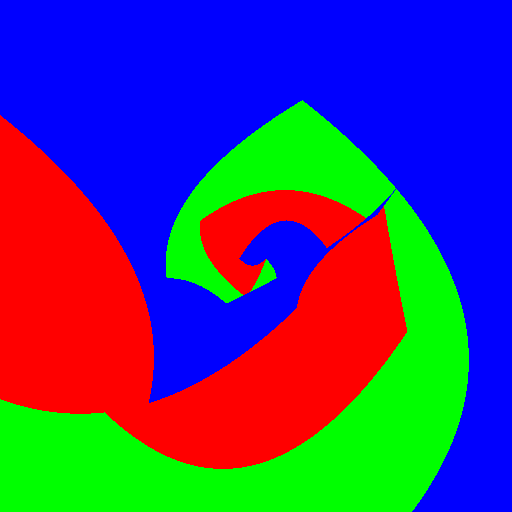
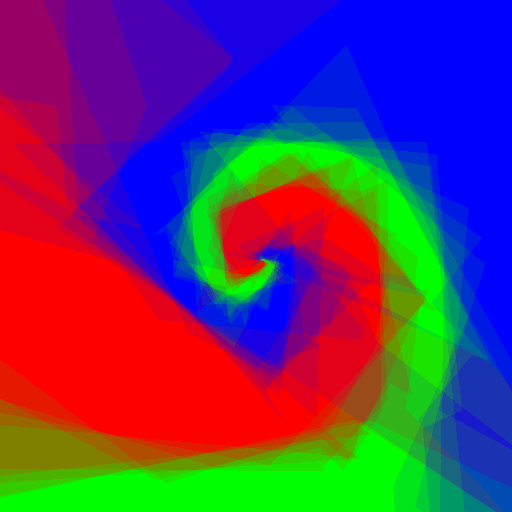
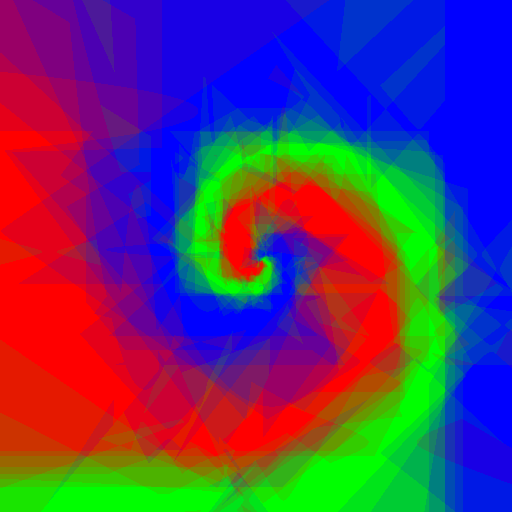
| Axis aligned | Linear | Conic | Parabola | |
|---|---|---|---|---|
| Tree | 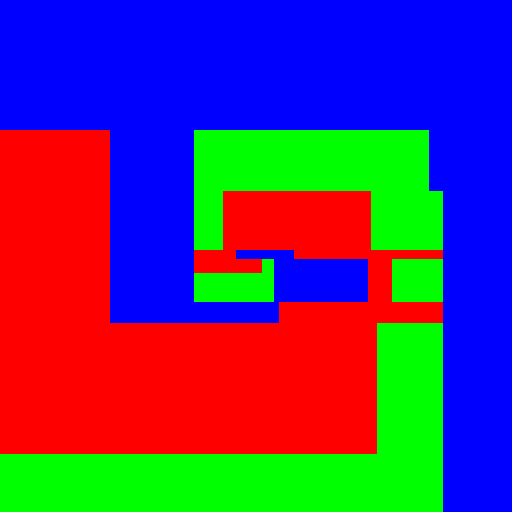 |
 |
 |
 |
| Forest |  |
 |
 |
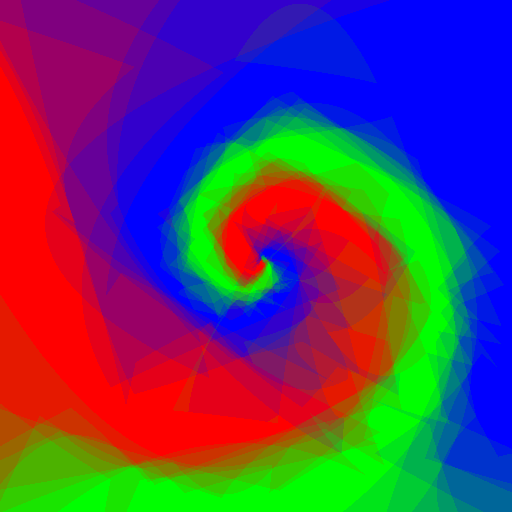 |
Experimenting with different weak learners in Random Forests.
Code adjustments:
- I switched to pillow to write images, which is known to work with Pypy, instead of OpenCV or scikit-image which I normally use. Experimenting with these libraries will be for later.
- Slight issue with the conversion from a numpy array to a PIL image:
img = Image.fromarray(img)
produces:TypeError: must be string or read-only buffer, not ndarray
A quick workaround is to explicitly request a conversion from string:img = Image.fromstring('RGB',img.shape[:2], img.tostring()) -
Slight issue with numpy arrays:
TypeError: astype() got an unexpected keyword argument 'casting'
According to the developers, "casting" is a valid keyword, anyway you can easily work around it as well. -
numpy.RandomStateis needed with joblib so that each tree is grown with a different random seed (otherwise you end up with identical trees):AttributeError: 'module' object has no attribute '_RandomState__RandomState_ctor'
but it is now fixed in master.
Parameters:
512x512 image, maximum tree depth 10, forest of 10 trees, 100 random tested generated at each node in the case of the pure Python implementation, and precomputed for the whole forest in the case of scikit-learn. Precomputing features in scikit-learn amounts to transforming the input data to a higher dimensional space. The runtime correspond to evaluating the code with the four types of weak learners presented in the figure above.
Runtime:
Single tree
| CPython | 35.9s |
| Pypy | 12.4s |
That's a 2.9 speedup.
Forest of 10 trees
| CPython | 11min 5s |
| Pypy | 3min 5s |
That's a 3.5 speedup
Training time is only a few seconds, the high runtime is due to the fact that we are going through the 512x512 image, evaluating the forest at each pixel. I only used joblib for training, mainly to check the Pypy compatibility as it had little impact on the training time. We can compare those results with using scikit-learn and precomputed features.
scikit-learn
| naive scikit-learn | 23min |
| numpy + scikit-learn | 5.2s |
In the first case, we extract features and do the classification pixel by pixel. This is what you should never do. Instead, use matrix operations and predict the whole image in one go. This is actually the approach I presented in my talk, but for a 3D volume you can also split the whole image in batches so that the extracted features fit in memory.
Conclusion:
If I can run some code 3 times faster with minor changes, I'll take it, especially if it makes Python's for loop bearable. Before going into code optimisation, it might be worth checking if Pypy can run it. After all, it is just a matter of calling a different binary. Hopefully, as time goes by, more and more packages will be compatible, but this also depends on its community growing. Thanks a lot to Romain and PyData for introducing me to Pypy, and hopefully next year there will be a benchmark CPython versus Pypy evaluating more complex image processing and machine learning tasks, and of course, running in a single notebook.



Thanks for giving PyPy a try, and helping us work through the issues you found. I just wanted to note that the casting keyword is supported in astype() on our post-2.6.0 nightly builds, available from http://buildbot.pypy.org/nightly/trunk
ReplyDelete